The lab is supported by the National Science Foundation (CBET/CAREER, DMS/NIGMS),
Cecil H. and Ida Green Chair in Systems Biology Science, and UTD.
The laboratory invites applications for graduate students and postdoctoral associates.
Please email your cover letter and CV to bleris@utdallas.edu
Recent papers with summaries
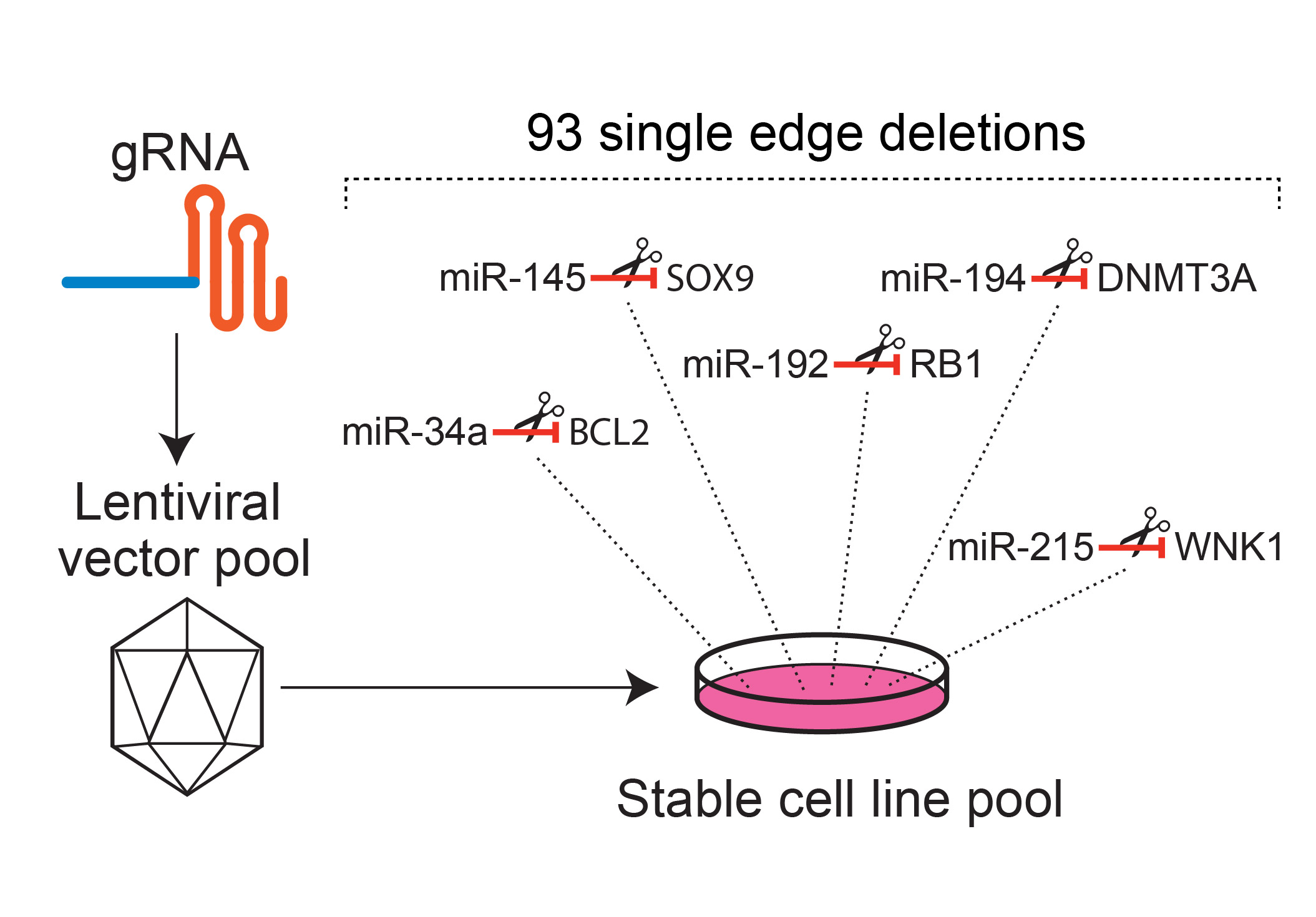
CRISPR-based editing reveals edge-specific effects in biological networks.
The CRISPR
Journal, 2018.
Networks consist of functional elements (nodes) that form a variety of diverse
connections (edges), with each node being a hub for multiple edges. Herein, in contrast
to node-centric network perturbation and analysis approaches, we present a
high-throughput CRISPR-based methodology for delineating the role of network edges.
|
|
|
|
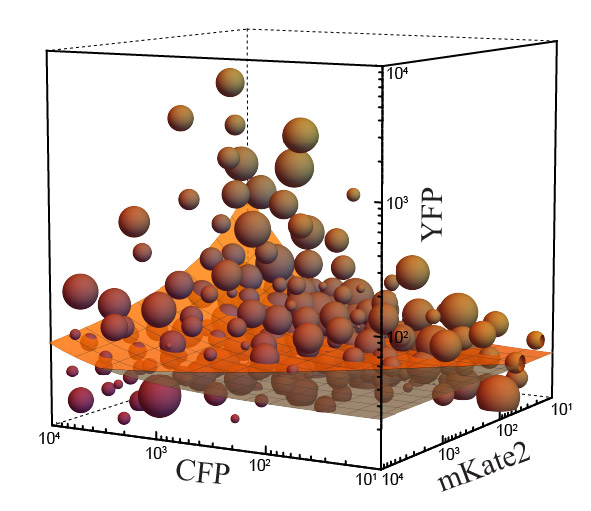
Mapping the operational landscape of microRNAs in synthetic gene circuits.
NPJ Systems Biology and
Applications, 2018.
We combined experiments and mathematical modeling to study the microRNA operational
landscape. We engineered custom genetic circuits that contain microRNA-based regulation
and introduced an analytical strategy, that includes clustering and superposition of
discrete experiments, to produce a “bird’s-eye view” perspective.
|
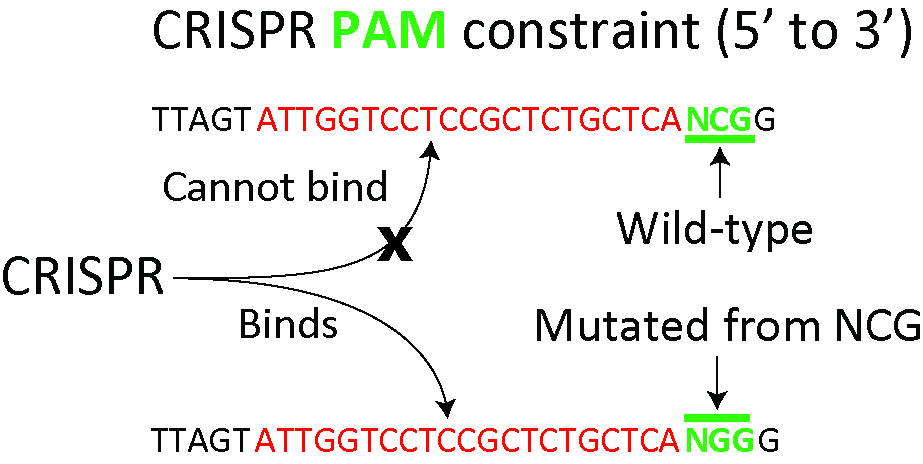
Exploiting the CRISPR/Cas9 PAM constraint for single-nucleotide resolution
interventions.
PLOS
ONE, 2016.
We demonstrate that the PAM requirement can be exploited to specifically target
single-nucleotide heterozygous mutations while exerting no aberrant effects on the
wild-type alleles. Specifically, we target the heterozygous G13A activating mutation of
KRAS in colorectal cancer cells and we show reversal of drug resistance to a MEK
small-molecule inhibitor.
|
|
|
|
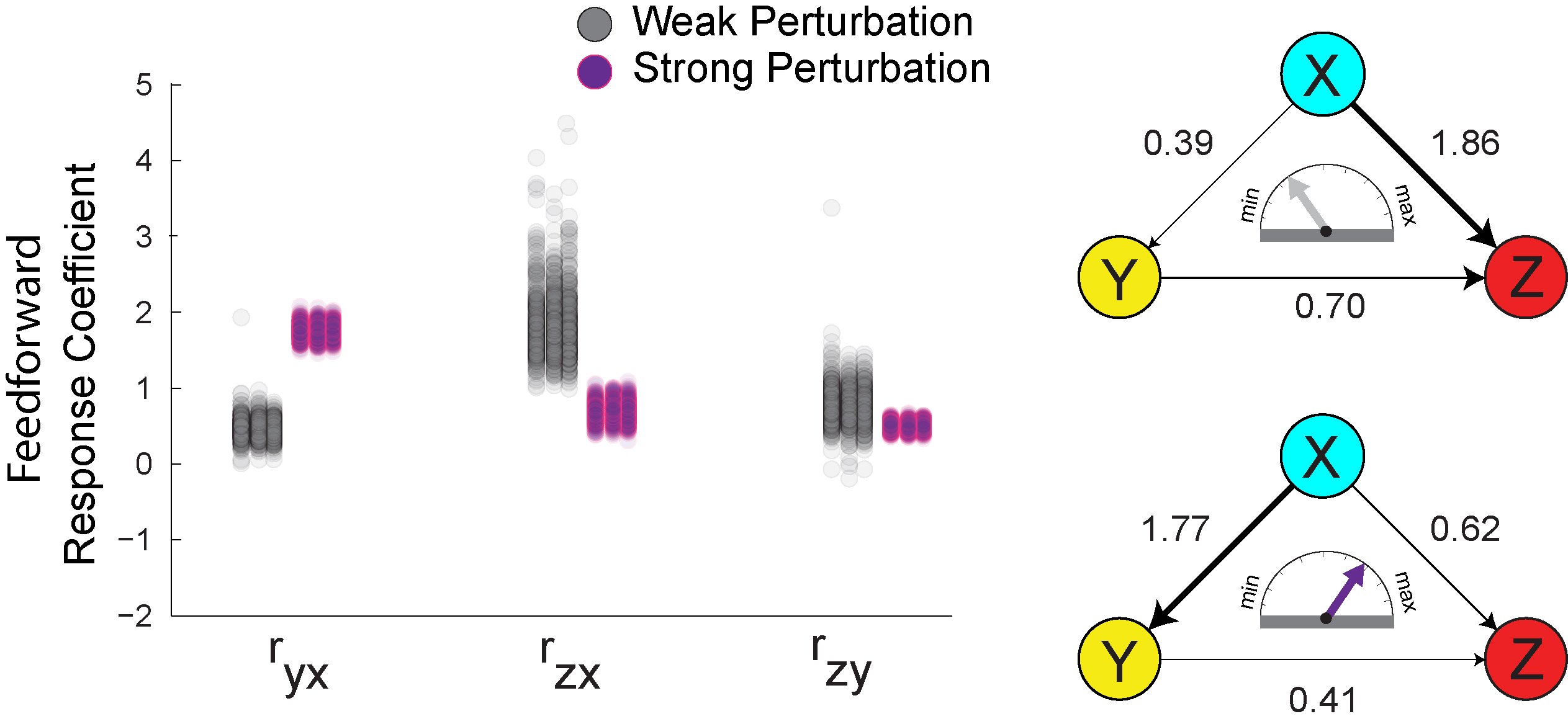
Discriminating direct and indirect connectivities in biological networks.
Proceedings
of the National Academy of Sciences (PNAS), 2015.
We used a combination of computational and theoretical approaches coupled to synthetic
biology experimentation in mammalian cells to study direct and indirect connectivities
in biological networks.
|
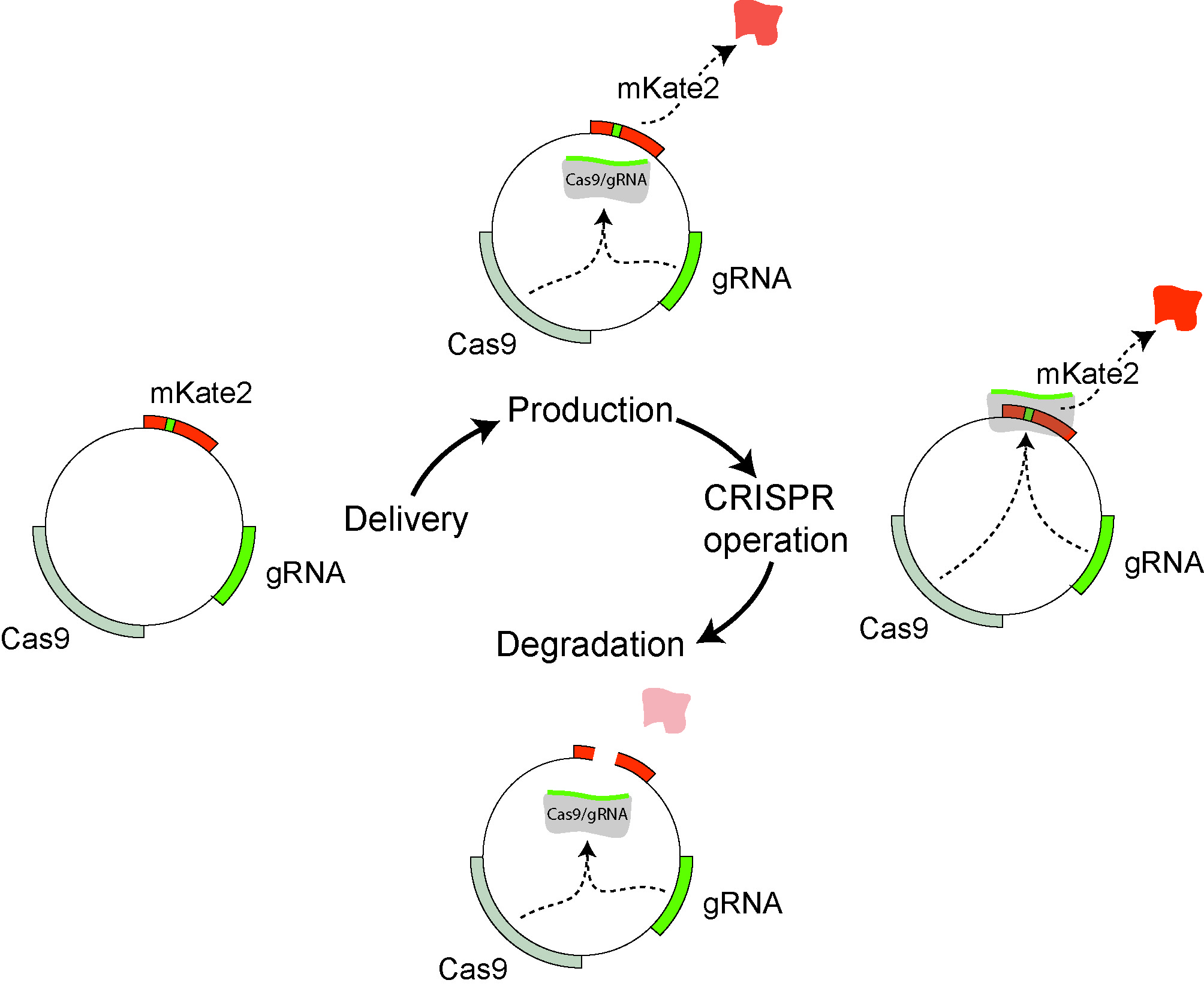
CRISPR-based self-cleaving mechanism for controllable gene delivery in human
cells.
Nucleic
Acids Research, 2015.
We introduce a methodology to control the copies and residence time of a gene product
delivered in host human cells but also selectively disrupt fragments of the delivery
vehicle. We envision future applications in complex synthetic biology architectures,
gene therapy and trace-free delivery.
|
|
|
|
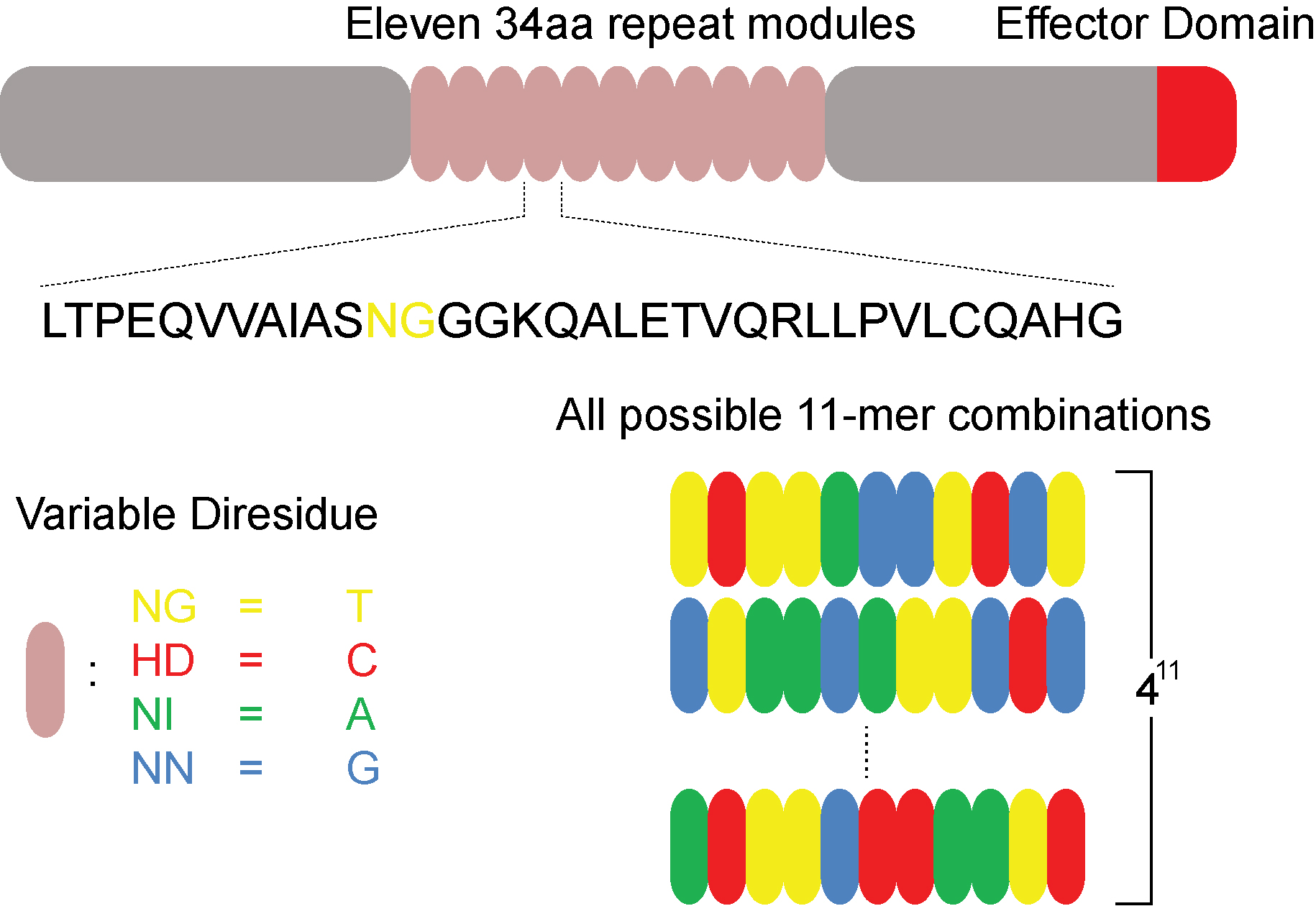
Assembly and validation of versatile transcription activator-like effector
libraries.
Scientific
Reports, 2014.
Here we introduce, describe the assembly, and demonstrate the use of comprehensive and
versatile transcription activator-like effector (TALE) libraries. Considering the highly
modular nature of TALEs and the versatility and ease of constructing these libraries we
envision broad implications for high-throughput genomic assays.
|
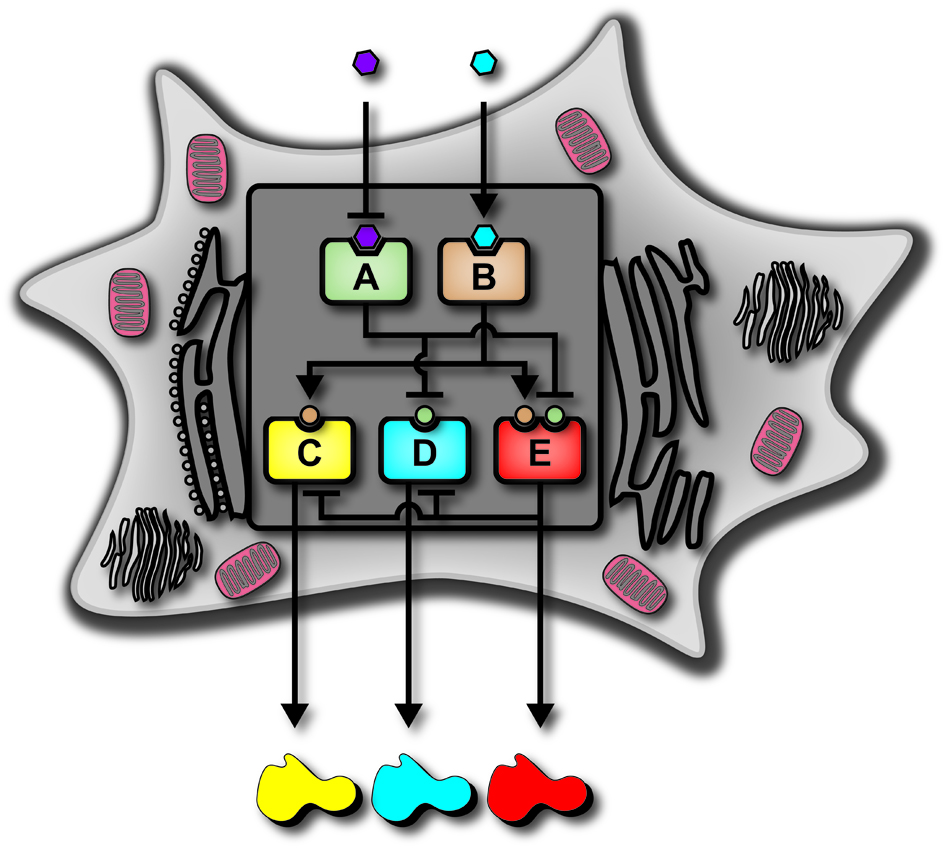
Biological 2-input decoder circuit in human cells.
ACS Synthetic Biology,
2014.
Decoders are combinational circuits that convert information from n inputs to a maximum
of 2n outputs. We present the first implementation of a genetic decoder in
human kidney cells.
|
|
|
|
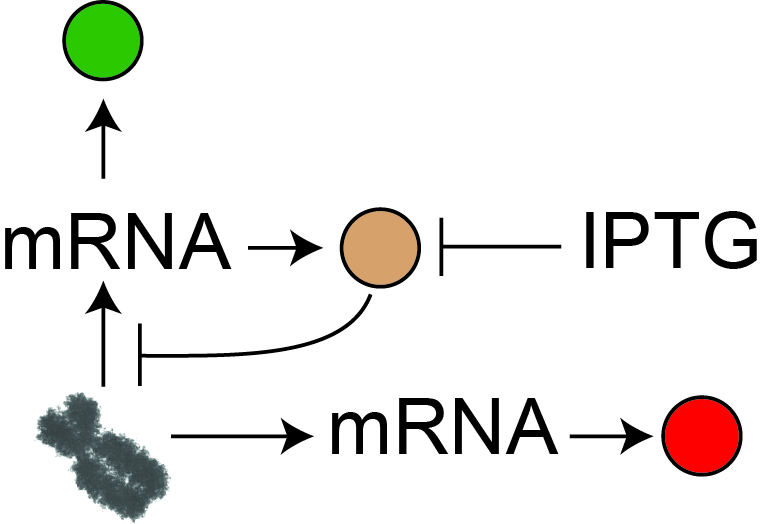
Synthetic mammalian transgene negative autoregulation.
Nature/EMBO Molecular Systems
Biology, 2013.
The effect of negative feedback on global and local sources of uncertainty is studied
with synthetic circuits stably integrated in human cells. Negative feedback is shown to
be the most efficient way to mitigate the effects of global fluctuations by introducing
a single additional regulatory link.
|
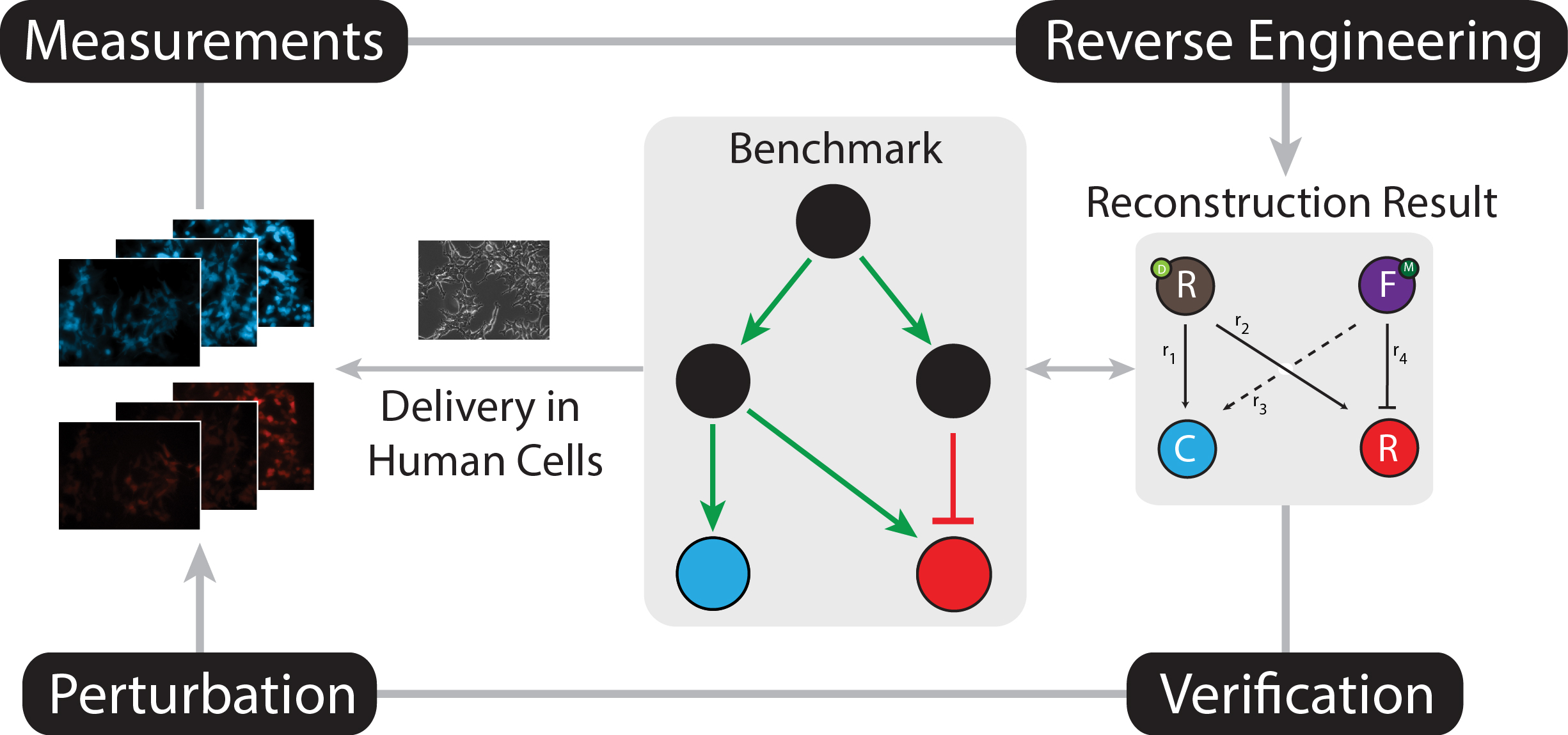
Reverse engineering validation using a benchmark synthetic gene circuit in human
cells.
ACS Synthetic Biology,
2013.
We stably integrate a synthetic gene network in human kidney cells and use it as a
benchmark for validating reverse engineering methodologies.
|
|









